Eukaryotes
Eukaryota, Organisms with nucleated cells
Patrick Keeling and Brian S. Leander

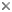
This tree diagram shows the relationships between several groups of organisms.
The root of the current tree connects the organisms featured in this tree to their containing group and the rest of the Tree of Life. The basal branching point in the tree represents the ancestor of the other groups in the tree. This ancestor diversified over time into several descendent subgroups, which are represented as internal nodes and terminal taxa to the right.

You can click on the root to travel down the Tree of Life all the way to the root of all Life, and you can click on the names of descendent subgroups to travel up the Tree of Life all the way to individual species.
For more information on ToL tree formatting, please see Interpreting the Tree or Classification. To learn more about phylogenetic trees, please visit our Phylogenetic Biology pages.
close boxThis tree is derived from a consensus of several different studies that are cited in the Discussion of Phylogenetic Relationships and the individual pages for each lineage shown.
Introduction
Even if you do not know the word ‘eukaryote’, you are already familiar with what they are, because you and nearly all other life forms that you experience with your unaided eyes are eukaryotes. The vast majority of eukaryotes that we knowingly interact with each day, mainly land plants and animals, are large – macroscopic – organisms, usually consisting of trillions of individual cells (Fig. 1). Even using our rather limited senses, we can immediately tell that macroscopic eukaryotes represent enormous diversity on many different levels. However, the true diversity of eukaryotes is far greater than ordinary experiences would lead you to appreciate; most of the many millions of eukaryotic species on Earth are hidden from view, because most eukaryotic life forms are microscopic (Fig. 2; also see the middle four cells in the title image bar). The diversity of these microbial eukaryotes must be discovered and explored with powerful equipment and techniques such as electron microscopy and molecular biology.







Fig. 1. Examples of multicellular macro-eukaryotes, namely animals and land plants. From left to right: a lion (© 2004 Tambako The Jaguar); a brown seaweed, Fucus (© Patrick Keeling); a fern fiddlehead (Sadleria cyatheoides, © 2004 Eric Schuettpelz); a seastar (Ophioderma rubicundum) resting on a coral ( © 1998 M. Benjamin Cowan, Ocean Images); a bee pollinating a flower (© Patrick Keeling); a green vine snake (Ahaetulla nasuta, © 2006 Jayanth Sharma). By the way, one of these pictures is actually not really an animal or plant, can you guess which one? Click here to find out.
Eukaryotes (also referred to as the Eukaryota or the Eukarya) comprise one of the three recognized domains of cellular life, the other two being the Archaea (or Archaebacteria) and the Eubacteria (or Bacteria) (Cavalier-Smith, 1998; Gogarten et al., 1989; Iwabe et al., 1989; Woese, 1987; Woese and Fox, 1977; Woese et al., 1990). Eukaryotes are distinguished from Archaea and Eubacteria in many different ways, but most importantly, the cells of eukaryotes display a much greater degree of structural organization and complexity. Archaeal and eubacterial cells generally lack internal structural organization (with a few notable exceptions, like the cyanobacteria). Eukaryotic cells, by contrast, share several complex structural characteristics. Most of these are parts of two interrelated systems: the cytoskeletal system and a system of membrane-delimited compartments. The cytoskeleton is an elaborate and highly organized internal scaffolding of proteins, such as actin-based microfilaments and tubulin-based microtubules. It also includes several molecular motors, such as kinesins and dyneins that provide the dynamic forces necessary for import and export mechanisms and many different modes of cell locomotion. Internal membrane-delimited compartments include mitochondria and plastids as well as different elements of the endomembrane system: the endoplasmic reticulum, Golgi bodies, vacuoles, and the nuclear envelope. The word ‘eu-karyote’ literally means ‘true kernel’, in reference to the sequestering of the genome into the membrane-bounded compartment called the nucleus.


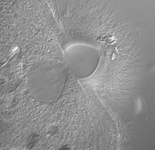
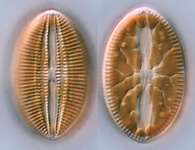

Fig. 2. Examples of micro-eukaryotes. From left to right: Selenidium serpulae, a gregarine apicomplexan parasite of polychaete worms (© Brian S. Leander); Eucomonympha imla, a parabasalian (© Patrick Keeling); two views of Diploneis, a diatom (© David G. Mann); Polykrikos lebourae, a dinoflagellate (© Mona Hoppenrath, Sarah Sparmann and Brian S. Leander); Trepomonas sp., a diplomonad (© Ivan Cepicka).
With these basic building blocks, eukaryotes have evolved an amazing array of structural and behavioral characters. One of the most significant innovations is the ability to engulf and internalize particles and other cells, a process called endocytosis or phagocytosis (literally meaning ‘cell eating’). This mode of nutrition opened up many new predatory niches that ultimately facilitated the formation of permanent associations between very different life forms via endosymbiosis (Stanier, 1970). Endosymbiotic associations have provided eukaryotes with much of their central metabolism, which has remained relatively conserved throughout the group's history. Overall, Archaea and Eubacteria show tremendous diversity in their metabolic capacities, but fairly limited morphological and behavioral diversity; conversely, eukaryotes share relatively similar (albeit sloppy) metabolic machinery but have undergone tremendous evolutionary diversification in morphology and behavior.
Characteristics
The known diversity of morphological characters in eukaryotes is simply staggering and can be attributed to the vast multitude of possible solutions to basic biological problems, such as nutrition/feeding, locomotion, defense, refuge, mate selection and reproduction. Eukaryotes are built from one or more internally differentiated cells comprised of intricate subcellular systems. Several single-celled lineages, for instance, have reached the utmost degree of morphological complexity within the confines of a single enveloping cell membrane (e.g. parabasalids, ciliates, dinoflagellates), while others have reached the lower limits of morphological complexity by becoming extremely streamlined (e.g. picophytoeukaryotes, yeasts). Moreover, some multicellular eukaryotes have struck the upper physical limits of overall body size (e.g. dinosaurs, elephants, and whales), while others are miniaturized to the point of being smaller than single-celled counterparts in the same ecosystem (e.g. gastrotrichs, tardigrades, rotifers and nematodes). Regardless of major differences in body size and morphological peculiarities, eukaryotes share many characteristics in common. Many of these characteristics are homologous for the entire group, whether comparing a blue whale to an amoeba or a human to a giant redwood tree.
Unifying Features of Eukaryotes
Below is a list of important features that are likely to have been present in the common ancestor of eukaryotes. Some of these features are still universally found in all eukaryotic diversity, while others have been lost or drastically transformed in some lineages, but are nevertheless ancestral to those groups (see Fig. 3 for examples).
- Cytoskeleton consisting of tubulin-based microtubules and actin-based microfilaments, and ancestrally including motile cell extensions called ‘flagella’ or ‘cilia’ that contain an axoneme of 9 peripheral microtubular doublets and 2 central microtubules.
- An endomembrane system that consists of endoplasmic reticulum, Golgi bodies, vacuoles, lysosomes, peroxisomes, and the nuclear envelope.
- Primary genome of each cell consisting of multiple linear chromosomes contained within a membrane-bound nucleus. Following replication of the genome the chromosomes are segregated by the process of mitosis. Cells in many species can have more than one nucleus.
- Mitochondria - organelles with diverse functions, usually including aerobic respiration, iron sulfur cluster assembly, and synthesis and breakdown of small molecules such as lipids and amino acids. Mitochondria are bounded by two membranes, and usually contain a small genome. They are the descendents of an alpha-proteobacterial endosymbiont.
- Translation machinery in the form of 80S ribosomes, each consisting of four molecules of RNA complexed with many proteins, and partitioned in a small (40S) and a large (60S) subunit.

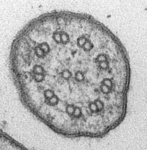
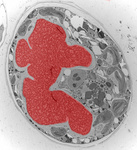
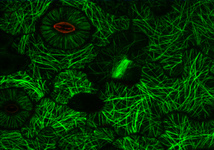
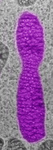
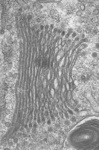
Fig. 3. Some unifying characteristics of eukaryotes. From left to right: the 9+2 microtubule structure of eukaryotic flagella (© Patrick Keeling and Kevin Carpenter), the nucleus (highlighted in red) from the diatom endosymbiont of the dinoflagellate Durinskia baltica (© Patrick Keeling and Kevin Carpenter), the microtubules of the Arabidopsis thaliana cytoskeleton highlighted by fluorescence (© Jessica Lucas and Fred Sack), a linear chromosome from a dinoflagellate nucleus (© Patrick Keeling and Kevin Carpenter), and the Golgi body from Microjoenia fallax (© Guy Brugerolle).
Other Common Characteristics of Eukaryotes
A number of other characteristics are common to many eukaryotes and not to prokaryotes, but these are not ancestral to all eukaryotes, and many have evolved several times independently (See Fig. 4 for examples).
- Multicellularity and tissue formation (e.g. green algae, land plants, red algae, brown algae, animals and fungi).
- Secreted hard parts (e.g. mollusk shells, plant cell walls, ecdysozoan cuticles, coccoliths, vertebrate endoskeletons, chrysophyte scales, polychaete tubes, diatom frustules, brachiopod shells, cnidarian corallites, euglenophyte loricas, poriferan spicules, echinoderm ossicles, foraminiferan and radiozoan tests).
- Extrusive organelles that function in defense, prey capture or parasitic invasion (e.g. ejectisomes of cryptomonads; trichocysts of alveolates; polar tubes of microsporidian fungi, gun cells of oomycetes; nematocysts of cnidarians, myxozoans and some dinoflagellates).
- Plastids, including chloroplasts and their homologues. Referring to plastids as homoplasies is a qualified statement, since the vast majority of plastids do ultimately stem from a common primary endosymbiosis with a cyanobacterium (the one possible exception being the ‘chromophore’ of the euglyphid amoeba Paulinella), but their subsequent spread via secondary and tertiary endosymbioses has led to a complicated distribution on the tree of eukaryotes (see Symbiosis section below).
Fig. 4. Some features are common, but not necessarily ancestral characteristics of eukaryotes. For example, making hard bones, shells, or other body parts. From left to right: the shell of the cephalopod Spirula spirula (© 1996 Richard E. Young), an SEM showing the external scales of the chrysophyte Mallomonas sp. (© 2003 Brian S. Leander), a collection of human bones from the Chapela dos Ossos in Évora, Portugal (© Patrick Keeling), and Calliarthron tuberculosum from the North East Pacific (© Patrick Keeling).
Role of Endosymbiosis in Eukaryotic Evolution
In addition to providing a significant nutritional mode, the advent of endocytosis in an ancestor of living eukaryotes also enabled a completely new way to generate cellular change and complexity: endosymbiosis. Put simply, endosymbiosis is the process by which one cell is taken up by another and retained internally, such that the two cells live together and integrate at some level, sometimes permanently. Endosymbiotic interactions have been common in eukaryotic evolution, and many such partnerships persist today (Margulis, 1981). In two cases, however, endosymbiotic events had far-reaching effects on the evolution of life: these are the origins of mitochondria and plastids (chloroplasts).
Mitochondria are generally known as the energy-generating powerhouses of eukaryotic cells, where oxidative phosphorylation and electron transport metabolism takes place (Reichert and Neupert, 2004). They are also involved in several other jobs such as oxidation of fatty acids, amino acid metabolism, and assembly of iron-sulfur clusters (Lill et al., 1999; Lill and Kispal, 2000). They are bounded by two membranes, the innermost of which is generally highly infolded to form ‘cristae’ that take characteristic shapes, either flat, tubes, or paddle-shapes (Fig. 5) (Taylor, 1978). The presence of mitochondria is an ancestral trait in eukaryotes (Roger, 1999; van der Giezen and Tovar, 2005; van der Giezen et al., 2005; Williams and Keeling, 2003), although in certain anaerobes and microaerophiles they have radically reduced or transformed functions: in some cases they are not involved in energy production at all (e.g., the ‘mitosomes’ of microsporidia, diplomonads, and archaemoebae, or ‘hydrogenosomes’ of parabasalia, some ciliates, and some chytrid fungi) (Embley, 2006; Müller, 1993; Tovar et al., 1999; van der Giezen et al., 2005; Williams and Keeling, 2003). Mitochondria can be traced back to a single endosymbiosis of an alpha-proteobacterium (Andersson and Kurland, 1999; Gray et al., 1999; Gray and Doolittle, 1982; Gray et al., 2004; Lang et al., 1999).

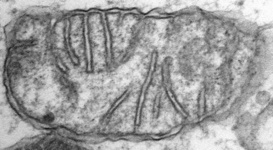
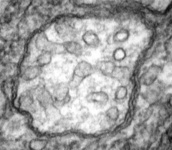
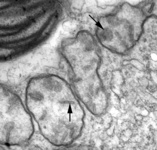
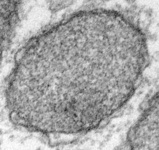
Fig. 5. Mitochondria. There are three main structural types of mitochondria, defined by the shape of their cristae, or infolding of the inner membrane: from left to right are flat cristae (from the animal Cryptocercus punctulatus, © Kevin Carpenter), tubular cristae (from the diatom endosymbiont of Kryptoperidinium foliaceum, © Patrick Keeling and Kevin Carpenter), discoidal cristae from the euglenid Eutreptia pertyi (© 2009 Brian S. Leander), and a hydrogenosome (with no cristae) from the parabasalian Trichonympha acuta (© Patrick Keeling and Kevin Carpenter).
Plastids are the photosynthetic organelles of plants and algae. “Plastid” is a general term for all such organelles, including chloroplasts (in the green lineage), rhodoplasts (in the red lineage), leucoplasts (colourless plastids), etc (Fig. 6). Plastids have diverse functions in addition to photosynthesis, including the biosynthesis of amino acids, fatty acids and isoprenoids (Harwood, 1996; Herrmann and Weaver, 1999; Rohdich et al., 2001). As in the case of mitochondria, plastids in many lineages have been radically reduced or transformed, primarily through the loss of photosynthesis (e.g., the ‘apicoplast’ of Apicomplexa, and the relict plastids of many parasitic algae and plants (Gould et al., 2008; Ralph et al., 2004; Wilson, 2002)). Plastids can also be traced back to a single endosymbiosis event involving a cyanobacterium and the ancestor of the Archaeplastida (Reyes-Prieto et al., 2007; Rodriguez-Ezpeleta et al., 2005). However, unlike mitochondria, plastids then spread to other eukaryotic lineages by secondary and tertiary endosymbiotic events (Archibald, 2005; Gould et al., 2008; Keeling, 2004; McFadden, 1999). In these events, one eukaryotic cell took up another eukaryote that already contained a plastid (an alga), and this second, endosymbiotic eukaryote was then reduced and integrated. In most cases all that remains of this alga is the plastid surrounded by the remains of the endosymbiont’s plasma membrane. However, in cryptomonads and chlorarachniophytes a tiny relict of the algal nucleus called a “nucleomorph” is also retained, the study of which helped elucidate the complex evolutionary history of plastids (Archibald, 2005; Douglas et al., 2001; Gilson et al., 2006; McFadden et al., 1997). Other endosymbiotic relationships based on photosynthesis are also known (Johnson et al., 2007; Okamoto and Inouye, 2005; Rumpho et al., 2008), but typically these are not integrated to the extent that they are generally accepted to be ‘organelles’ rather than ‘endosymbionts’. One possible exception is the euglyphid amoeba Paulinella chromatophora, where a cyanobacterium similar to Synechococcus or Prochlorococcus has been integrated to an extent approaching that of canonical plastids (Nowack et al., 2008).

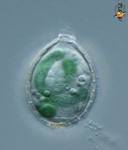
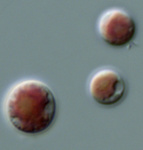

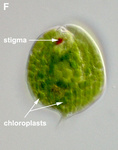
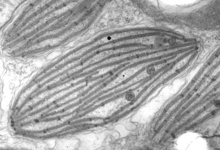
Fig. 6. Plastids. There are many different types of plastids, characterised by different pigments, structures, and envelopes. Here are a few examples, from left to right: the blue-green colored primary "chromophores" that were independently acquired by Paulinella chromatophora (© 2001 Michele Bahr and David Patterson), the red colored primary plastids of the red alga Porphyridium (© 2001 D. J. Patterson, L. Amaral-Zettler, M. Peglar and T. Nerad), the brown colored secondary plastids of Mallomonas insignis (© 2005 William Bourland), the green-colored secondary plastids of the euglenid Phacus (© Heather Esson and Brian S. Leander), and a TEM view of secondary green plastid of the euglenid Eutreptia pertyi (© Brian S. Leander).
Discussion of Phylogenetic Relationships
Our understanding of eukaryotic relationships has been transformed by the use of molecular data to reconstruct phylogenies (Sogin et al., 1986). Prior to that, the diversity of microbial eukaryotes was vastly underestimated, and the relationships between them and multicellular eukaryotes were difficult to resolve (Taylor, 1978). Early molecular phylogenies based on small subunit ribosomal RNA (SSU rRNA) gene sequences suggested a ladder of basal lineages topped by a ‘crown’ composed of multicellular groups (animals, plants, and fungi) together with a subset of the purely microbial lineages (Sogin, 1989). A great number of the relationships revealed by SSU rRNA phylogeny have stood the test of time, but subsequent analyses based on protein coding genes and more recently very large datasets composed of hundreds of protein coding genes have led to a revision of the overall structure of the tree. The current view of eukaryotic phylogeny is of a small number of large ‘supergroups’, each comprising a spectacular diversity of structures, nutritional modes, and behaviours (Adl et al., 2005; Keeling, 2004; Keeling et al., 2005; Simpson and Roger, 2002). Some of these supergroup hypotheses are well supported, while others remain the subject of vigorous debate (see (Keeling et al., 2005) for a discussion of evidence). Furthermore the relationships between supergroups are poorly understood. Below we summarise the main members of each supergroup, the evidence for its monophyly, and emerging hypotheses for inter-supergroup relationships.
Archaeplastida (Plantae)
The Archaeplastida, or Plantae, comprises glaucophytes, red algae, green algae and plants. They are united by the possession of a plastid derived from primary endosymbiosis (see Symbiosis section). There has long been strong support for the monophyly of plastids in Archaeplastida based on molecular phylogeny and also plastid genome structure (Turner, 1997; Turner et al., 1999), and molecular phylogenies based on large numbers of protein coding genes have more recently demonstrated the monophyly of the nuclear/cytosolic lineage as well (Burki et al., 2008; Moreira et al., 2000; Reyes-Prieto et al., 2007).


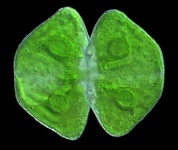


Fig. 7. Some examples of Archaeplastida. From left to right, the red alga Chondracanthus (© Patrick Keeling), the green alga Cosmarium (© Patrick Keeling) and the land plants Typha latifolia (© Patrick Keeling) and Calocedrus decurrens (© 2008 Fort Photo).
Excavata
Excavata is a large and diverse grouping that has been proposed based on a synthesis of morphological and molecular data. Many excavates share a similar feeding groove structure (from which the name is derived) (Simpson and Patterson, 2001; Simpson and Patterson, 1999). Many others lack this structure, but are demonstrably related to lineages that possess it in molecular phylogenies (Simpson, 2003; Simpson et al., 2006; Simpson et al., 2002). Putting this evidence together led to the suggestion of shared ancestry, and some recent multi-gene phylogenies in fact provide tentative support for the monophyly of the whole group (Burki et al., 2008; Rodriguez-Ezpeleta et al., 2007). Many excavates are anaerobes/microaerophiles and contain mitosomes or hydrogenosomes (e.g. diplomonads and parabasalids). Some are important parasites of animals (e.g. trypanosomes, Giardia). One lineage, the euglenids, includes photosynthetic species that have plastids derived from a green alga by secondary endosymbiosis (Breglia et al., 2007; Leander et al., 2007).





Fig. 8. Some examples of Excavata. From left to right: an SEM of the oxymonad Saccinobaculus minor (© Kevin Carpenter and Patrick Keeling), an SEM of the photosynthetic euglenid Lepocinclis spirogyra (© 2003 Brian S. Leander), a DIC light micrograph of the heterolobosean Stephanopogon minuta (© Naoji Yubuki and Brian S. Leander) and a fluorescence micrograph of the parabasalid Holomastigotes elongatum (© Guy Brugerolle).
Chromalveolata
Chromalveolates comprises six major groups of primarily single celled eukaryotes: apicomplexans, dinoflagellates and ciliates are members of the alveolates, they are hypothesised to be related to stramenopiles, cryptomonads, and haptophytes (Cavalier-Smith, 2004; Keeling, 2009). The basis for this hypothesis is the widespread presence of plastids in these groups that are all derived from secondary endosymbiosis with a red alga. It was therefore proposed that all chromalveolates share a common ancestor where this endosymbiosis took place (Cavalier-Smith, 1999). The monophyly of the plastids has been demonstrated with limited sampling (Hagopian et al., 2004; Rogers et al., 2007; Yoon et al., 2002), and some phylogenies inferred from many different nuclear genes show that the Chromalveolata are monophyletic with the Rhizaria nested within (see below) (Hackett et al., 2007). Additional support comes from two genes with unusual evolutionary histories involving lateral gene transfer and/or re-targeting to the plastid that are most consistent with a common origin of chromalveolate plastids (Fast et al., 2001; Harper and Keeling, 2003; Patron et al., 2004).

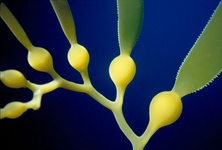

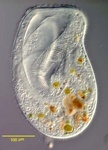

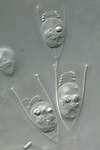
Fig. 9. Some examples of chromalveolates. From left to right: the multiculluar brown alga Macrocystis (© Tom Gruber), the diatom Stephanodiscus (© David G. Mann; this image is derived from the Professor Frank Round Image Archive at the Royal Botanic Garden Edinburgh), the ciliate Bursaria truncatella (© William Bourland, Freshwater and terrestrial microbes of Idaho), the filamentous dinoflagellate Gymnodinium catenatum (© Bob Andersen and D. J. Patterson; this image is of material from Provasoli-Guillard National Center for Culture of Marine Phytoplankton), and the bicosoecid Bicosoeca petiolata (© William Bourland, Freshwater and terrestrial microbes of Idaho).
Rhizaria
Rhizaria comprises several very large and diverse groups of amoebae, flagellates and amoeboflagellates (Cavalier-Smith and Chao, 2003). Many of these will not be familiar to many readers, but they are ubiquitous in nature and important predators in many environments. Major lineages include Cercozoa, Foraminifera, and Radiolaria. Rhizaria is the most recently recognized supergroup, having been identified exclusively from molecular phylogenetic reconstruction (Cavalier-Smith, 2002; Cavalier-Smith, 2003; Nikolaev et al., 2004). Prior to this, there was little reason to anticipate this grouping, because there is no major structural character that unites them. (Although the amoeboid members of the group tend to produce fine pseudopodia, rather than the broad pseudopodia seen in many Amoebozoa – see below.) However, analyses of molecular phylogenies based on nearly all genes examined, as well as rare molecular markers such as insertions and deletions, initially identified the Cercozoa as a group that has then expanded to include the Foraminifera and eventually the Radiolaria (Archibald et al., 2002; Bass et al., 2005; Burki et al., 2007; Burki et al., 2008; Keeling, 2001; Longet et al., 2003; Moreira et al., 2007; Nikolaev et al., 2004; Polet et al., 2004). Analyses of multiple protein coding genes have further supported the monophyly of Rhizaria, and suggested a relationship to chromalveolates (see below).

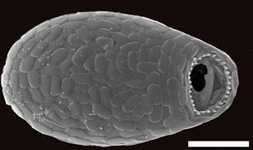
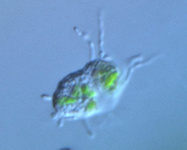
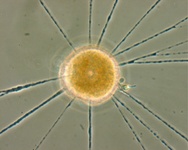
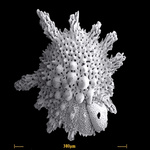
Fig. 10. Some examples of Rhizaria. From left to right: the euglyphid amoeba Corythion dubium (© Edward Mitchell), the chlorarachniophyte Gymnochlora (© Patrick Keeling), the foraminiferans Allogromia (© Jan Pawlowski and José Fahrni) and Calcarina spengleri (© 2008 Michael).
Opisthokonta
Opisthokonta is a grouping consisting of Animals (Metazoa), the true Fungi and their close protistan relatives. The closest relatives of animals include choanoflagellates, which are free-living unicellular or colonial flagellates, and the parasitic Ichthyosporea (also known as Mesomycetozoea). Fungi are most closely related to a group of amoebae called nucleariids. Opisthokonts share two conspicuous features that are uncommon in other eukaryotes: Almost all cells in this group have flat mitochondrial cristae, while flagellated cells typically have a single emergent flagellum that inserts at the posterior end of the cell (Cavalier-Smith, 1987). The monophyly of this group has been shown convincingly by molecular phylogenies (Baldauf and Palmer, 1993; Lang et al., 1999; Ragan et al., 1996; Ruiz-Trillo et al., 2006; Steenkamp et al., 2006; Wainright et al., 1993), and also by a large, conserved insertion within the protein Elongation Factor 1-alpha (Baldauf and Palmer, 1993; Steenkamp et al., 2006). Recently a possible shared lateral gene transfer has been reported (Huang et al., 2005).

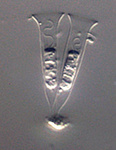



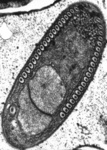
Fig. 11. Some examples of opisthokonts. From left to right: the choanoflagellate Salpingoeca (© William Bourland), the animals Saimiri sciureus (squirrel monkey, © Luc Viatour) and Cyanea capillata (lion's mane sea jelly, © Patrick Keeling), the fungus Morchella (© Patrick Keeling), and the microsporidian Fibrillanosema crangonycis (© Leon White).
Amoebozoa
The Amoebozoa are a diverse collection of protozoan eukaryotes, almost all of which are amoebae (i.e. cells that produce pseudopodia, but lack flagella) for some or all of their life cycle. Many produce lobose or fan-shaped pseudopodia (in contrast to the elongate, fine pseudopodia typical of Rhizaria), although short, fine sub-pseudopodia are also common. Amoebozoa includes lineages of ‘lobose amoebae’ (e.g the well known Amoeba and Chaos), the lobose testate amoebae (with the cell enclosed in a shell), most of the lineages of ‘slime molds’, the pelobionts and Entamoebae, which lack classical mitochondria, and a few mitochondriate flagellates. Amoebozoa were only recently united as group. Detailed microscopy studies had shown that amoebae as a whole were polyphyletic, and thus when early molecular phylogenetic studies based especially on ribosomal RNA sequences placed slime molds, lobose amoebae, pelobionts and entamoebae as multiple independent lineages (Hinkle et al., 1994; Sogin, 1989), this result seemed plausible. In the last few years, increasingly sophisticated molecular phylogenies incorporating many more taxa and/or genes have tended to unite these previously disparate groups (Bapteste et al., 2002; Fahrni et al., 2003), though not always with strong statistical support. A recent study suggests that the pseudopodia-producing flagellate Breviata represents the deepest branch within a monophyletic amoebozoa clade (Minge et al., 2008).



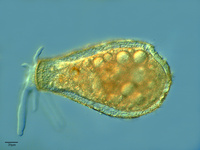
Fig. 12. Some examples of amoebozoans. From left to right: the slime mold Leocarpus fragilis (© 2008 Mavi Rodriguez Garcia), the slime mold Diachea leucopodia (© 2006 Randy Darrah, The Eumycetozoan Project, University of Arkansas), and a lobosean testate amoeba, likely Nebela tubulosa (© 2009 Antonio Guillén, identified by Ralf Meisterfeld).
'Unikonts': A Clade Consisting of Opisthokonts & Amoebozoans
There is now considerable evidence from molecular phylogenies that the opisthokonts and amoebozoans are closely related (Baldauf et al., 2000; Bapteste et al., 2002), and they also share a handful of other molecular characteristics in common (Richards and Cavalier-Smith, 2005). They have been proposed to be a clade called ‘unikonts’ because many of these organisms have a single flagellum (Cavalier-Smith, 2002), but biflagellated lineages are also known in this group. The root of the tree of eukaryotes has been proposed to be somewhere near this lineage, so it is possible the ‘unikonts’ are paraphyletic (Stechmann and Cavalier-Smith, 2002; Stechmann and Cavalier-Smith, 2003).
Do rhizarians branch within the chromalveolates?
There has long been very strong evidence from several kinds of data for the monophyly of alveolates. Multi-gene trees have also consistently and strongly supported a relationship between alveolates and stramenopiles (Burki et al., 2007; Burki et al., 2008; Hackett et al., 2007; Patron et al., 2007; Rodriguez-Ezpeleta et al., 2005; Rodriguez-Ezpeleta et al., 2007; Simpson et al., 2006). There is now also very strong evidence from molecular phylogenies and a shared lateral gene transfer for the monophyly of cryptomonads, haptophytes, and their relatives (Burki et al., 2008; Hackett et al., 2007; Patron et al., 2007; Rice and Palmer, 2006). In addition there is evidence from the plastid genome and plastid targeted proteins for the monophyly of chromalveolates and their plastids (Fast et al., 2001; Hagopian et al., 2004; Harper and Keeling, 2003; Patron et al., 2004; Rogers et al., 2007; Yoon et al., 2002). However, multi-gene trees also consistently show that the entire rhizarian supergroup is closely related to alveolates and stramenopiles (Burki et al., 2007; Burki et al., 2008; Hackett et al., 2007; Rodriguez-Ezpeleta et al., 2007), and some support the monophyly of chromalveolates as a whole with the Rhizaria nested within the group. These relationships will doubtless be refined with further data, but for now we follow the consensus of the available evidence and place the Rhizaria within the Chromalveolata.
References
For additional references about eukaryote phylogeny and evolution please see Eukaryotes - Comprehensive List of References.
Adl, S.M., Simpson, A.G., Farmer, M.A., Andersen, R.A., Anderson, O.R., Barta, J.R., Bowser, S.S., Brugerolle, G., Fensome, R.A., Fredericq, S., James, T.Y., Karpov, S., Kugrens, P., Krug, J., Lane, C.E., Lewis, L.A., Lodge, J., Lynn, D.H., Mann, D.G., McCourt, R.M., Mendoza, L., Moestrup, O., Mozley-Standridge, S.E., Nerad, T.A., Shearer, C.A., Smirnov, A.V., Spiegel, F.W. and Taylor, M.F. (2005) The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol., 52, 399-451.
Andersson, S.G. and Kurland, C.G. (1999) Origins of mitochondria and hydrogenosomes. Curr. Opin. Microbiol., 2, 535-541.
Archibald, J.M. (2005) Jumping genes and shrinking genomes--probing the evolution of eukaryotic photosynthesis with genomics. IUBMB Life, 57, 539-547.
Archibald, J.M., Longet, D., Pawlowski, J. and Keeling, P.J. (2002) A novel polyubiquitin structure in Cercozoa and Foraminifera: evidence for a new eukaryotic supergroup. Mol. Biol. Evol., 20, 62-66.
Baldauf, S.L. and Palmer, J.D. (1993) Animals and fungi are each other's closest relatives: congruent evidence from multiple proteins. Proc. Natl. Acad. Sci. USA, 90, 11558-11562.
Baldauf, S.L., Roger, A.J., Wenk-Siefert, I. and Doolittle, W.F. (2000) A kingdom-level phylogeny of eukaryotes based on combined protein data. Science, 290, 972-977.
Bapteste, E., Brinkmann, H., Lee, J., Moore, D., Sensen, C., Gordon, P., Durufle, L., Gaasterland, T., Lopez, P., Muller, M. and Philippe, H. (2002) The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostellium, Entamoeba, and Mastigamoeba. Proc. Natl. Acad. Sci. U S A, 99, 1414-1419.
Bass, D., Moreira, D., Lopez-Garcia, P., Polet, S., Chao, E.E., von der Heyden, S., Pawlowski, J. and Cavalier-Smith, T. (2005) Polyubiquitin insertions and the phylogeny of Cercozoa and Rhizaria. Protist, 156, 149-161.
Breglia, S.A., Slamovits, C.H. and Leander, B.S. (2007) Phylogeny of phagotrophic euglenids (Euglenozoa) as inferred from hsp90 gene sequences. J. Eukaryot. Microbiol., 54, 86-92.
Burki, F., Shalchian-Tabrizi, K., Minge, M., Skjaeveland, A., Nikolaev, S.I., Jakobsen, K.S. and Pawlowski, J. (2007) Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE, 2, e790.
Burki, F., Shalchian-Tabrizi, K. and Pawlowski, J. (2008) Phylogenomics reveals a new 'megagroup' including most photosynthetic eukaryotes. Biol. Lett., 4(4), 366-369.
Cavalier-Smith, T. (1987) The origin of fungi and pseudofungi. In Rayner, A.D.M., Brasier, C.M. and Moore, D. (eds.), Evolutionary biology of the fungi. Cambridge University Press, Cambridge, pp. 339-353.
Cavalier-Smith, T. (1998) A revised six-kingdom system of life. Biol. Rev. Camb. Philos. Soc., 73, 203-266.
Cavalier-Smith, T. (1999) Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J. Eukaryot. Microbiol., 46, 347-366.
Cavalier-Smith, T. (2002) The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Sys. Evol. Microbiol., 52, 297-354.
Cavalier-Smith, T. (2003) Protist phylogeny and the high-level classification of Protozoa. Eur. J. Protistol., 39, 338-348.
Cavalier-Smith, T. (2004) Chromalveolate diversity and cell megaevolution: interplay of membranes, genomes and cytoskeleton. In Hirt, R.P. and Horner, D. (eds.), Organelles, Genomes and Eukaryotic Evolution. Taylor and Francis, London, pp. 71-103.
Cavalier-Smith, T. and Chao, E.E. (2003) Phylogeny and classification of phylum Cercozoa (Protozoa). Protist, 154, 341-358.
Douglas, S., Zauner, S., Fraunholz, M., Beaton, M., Penny, S., Deng, L.T., Wu, X., Reith, M., Cavalier-Smith, T. and Maier, U.G. (2001) The highly reduced genome of an enslaved algal nucleus. Nature, 410, 1091-1016.
Embley, T.M. (2006) Multiple secondary origins of the anaerobic lifestyle in eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci., 361, 1055-1067.
Fahrni, J.F., Bolivar, I., Berney, C., Nassonova, E., Smirnov, A. and Pawlowski, J. (2003) Phylogeny of lobose amoebae based on actin and small-subunit ribosomal RNA genes. Mol. Biol. Evol., 20, 1881-1886.
Fast, N.M., Kissinger, J.C., Roos, D.S. and Keeling, P.J. (2001) Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol. Biol. Evol., 18, 418-426.
Gilson, P.R., Su, V., Slamovits, C.H., Reith, M.E., Keeling, P.J. and McFadden, G.I. (2006) Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature's smallest nucleus. Proc. Natl. Acad. Sci. USA, 103, 9566-9571.
Gogarten, J.P., Kiblak, H., Dittrich, P., Taiz, L., Bowman, E.J., Bowman, B.J., Manolson, N.F., Poole, R.J., Date, T., Oshima, T., Konishi, J., Denda, K. and Yoshida, M. (1989) Evolution of the vacuolar H+-ATPase: inplications for the origin of eukaryotes. Proc. Natl. Acad. Sci. USA, 86, 6661-6665.
Gould, S.B., Waller, R.F. and McFadden, G.I. (2008) Plastid evolution. Annu. Rev. Plant. Biol., 59, 491-517.
Gray, M.W., Burger, G. and Lang, B.F. (1999) Mitochondrial evolution. Science, 283, 1476-1481.
Gray, M.W. and Doolittle, W.F. (1982) Has the endosymbiont hypothesis been proven? Microbiol Rev., 46, 1-42.
Gray, M.W., Lang, B.F. and Burger, G. (2004) Mitochondria of protists. Annu. Rev. Genet., 38, 477-524.
Hackett, J.D., Yoon, H.S., Li, S., Reyes-Prieto, A., Rummele, S.E. and Bhattacharya, D. (2007) Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates. Mol. Biol. Evol., 24, 1702-1713.
Hagopian, J.C., Reis, M., Kitajima, J.P., Bhattacharya, D. and de Oliveira, M.C. (2004) Comparative analysis of the complete plastid genome sequence of the red alga Gracilaria tenuistipitata var. liui provides insights into the evolution of rhodoplasts and their relationship to other plastids. J. Mol. Evol., 59, 464-477.
Harper, J.T. and Keeling, P.J. (2003) Nucleus-encoded, plastid-targeted glyceraldehyde-3-phosphate dehydrogenase (GAPDH) indicates a single origin for chromalveolate plastids. Mol. Biol. Evol., 20, 1730-1735.
Harwood, J.L. (1996) Recent advances in the biosynthesis of plant fatty acids. Biochim. Biophys. Acta, 1301, 7-56.
Herrmann, K.M. and Weaver, L.M. (1999) The Shikimate Pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol., 50, 473-503.
Hinkle, G., Leipe, D.D., Nerad, T.A. and Sogin, M.L. (1994) The unusually long small subunit ribosomal RNA of Phreatamoeba balamuthi. Nucleic Acids Res., 22, 465-469.
Huang, J., Xu, Y. and Gogarten, J.P. (2005) The presence of a haloarchaeal type tyrosyl-tRNA synthetase marks the opisthokonts as monophyletic. Mol. Biol. Evol., 22, 2142-2146.
Iwabe, N., Kuma, K.-I., Hasegawa, M., Osawa, S. and Miyata, T. (1989) Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc. Natl. Acad. Sci. USA, 86, 9355-9359.
Johnson, M.D., Oldach, D., Delwiche, C.F. and Stoecker, D.K. (2007) Retention of transcriptionally active cryptophyte nuclei by the ciliate Myrionecta rubra. Nature, 445, 426-428.
Keeling, P.J. (2001) Foraminifera and Cercozoa are related in actin phylogeny: two orphans find a home? Mol. Biol. Evol., 18, 1551-1557.
Keeling, P.J. (2004) The diversity and evolutionary history of plastids and their hosts. Am. J. Bot., 91, 1481-1493.
Keeling, P.J. (2009) Chromalveolates and the evolution of plastids by secondary endosymbiosis. J. Eukaryot Microbiol., in press.
Keeling, P.J., Burger, G., Durnford, D.G., Lang, B.F., Lee, R.W., Pearlman, R.E., Roger, A.J. and Gray, M.W. (2005) The tree of eukaryotes. Trends Ecol. Evol., 20, 670-676.
Lang, B.F., Gray, M.W. and Burger, G. (1999) Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet., 33, 351-397.
Leander, B.S., Esson, H.J. and Breglia, S.A. (2007) Macroevolution of complex cytoskeletal systems in euglenids. Bioessays, 29, 987-1000.
Lill, R., Diekert, K., Kaut, A., Lange, H., Pelzer, W., Prohl, C. and Kispal, G. (1999) The essential role of mitochondria in the biogenesis of cellular iron-sulfur proteins. Biol. Chem., 380, 1157-1166.
Lill, R. and Kispal, G. (2000) Maturation of cellular Fe-S proteins: an essential function of mitochondria. Trends Biochem. Sci., 25, 352-356.
Longet, D., Archibald, J.M., Keeling, P.J. and Pawlowski, J. (2003) Foraminifera and Cercozoa share a common origin according to RNA polymerase II phylogenies. Int. J. Syst. Evol. Microbiol., 53, 1735 - 1739.
Margulis, L. (1981) Symbiosis in cell evolution. W. H. Freeman and Co., San Francisco.
McFadden, G.I. (1999) Endosymbiosis and evolution of the plant cell. Curr. Opin. Plant. Biol., 2, 513-519.
McFadden, G.I., Gilson, P.R., Douglas, S.E., Cavalier-Smith, T., Hofmann, C.J. and Maier, U.G. (1997) Bonsai genomics: sequencing the smallest eukaryotic genomes. Trends Genet., 13, 46-49.
Minge, M.A., Silberman, J.D., Orr, R.J., Cavalier-Smith, T., Shalchian-Tabrizi, K., Burki, F., Skjaeveland, A. and Jakobsen, K.S. (2008) Evolutionary position of breviate amoebae and the primary eukaryote divergence. Proc. Biol. Sci., 276, 597-604.
Moreira, D., Le Guyader, H. and Phillippe, H. (2000) The origin of red algae and the evolution of chloroplasts. Nature, 405, 69-72.
Moreira, D., von der Heyden, S., Bass, D., Lopez-Garcia, P., Chao, E. and Cavalier-Smith, T. (2007) Global eukaryote phylogeny: Combined small- and large-subunit ribosomal DNA trees support monophyly of Rhizaria, Retaria and Excavata. Mol. Phylogenet. Evol., 44, 255-266.
Müller, M. (1993) The hydrogenosome. J. Gen. Microbiol., 139, 2879-2889.
Nikolaev, S.I., Berney, C., Fahrni, J.F., Bolivar, I., Polet, S., Mylnikov, A.P., Aleshin, V.V., Petrov, N.B. and Pawlowski, J. (2004) The twilight of Heliozoa and rise of Rhizaria, an emerging supergroup of amoeboid eukaryotes. Proc. Natl. Acad. Sci. USA, 101, 8066-8071.
Nowack, E.C., Melkonian, M. and Glockner, G. (2008) Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol., 18, 410-418.
Okamoto, N. and Inouye, I. (2005) A secondary symbiosis in progress? Science, 310, 287.
Patron, N.J., Inagaki, Y. and Keeling, P.J. (2007) Multiple gene phylogenies support the monophyly of cryptomonad and haptophyte host lineages. Curr. Biol., 17, 887-891.
Patron, N.J., Rogers, M.B. and Keeling, P.J. (2004) Gene replacement of fructose-1,6-bisphosphate aldolase (FBA) supports a single photosynthetic ancestor of chromalveolates. Eukaryot. Cell, 3, 1169-1175.
Polet, S., Berney, C., Fahrni, J. and Pawlowski, J. (2004) Small-subunit ribosomal RNA gene sequences of Phaeodarea challenge the monophyly of Haeckel's Radiolaria. Protist, 155, 53-63.
Ragan, M.A., Goggin, C.L., Cawthorn, R.J., Cerenius, L., Jamieson, A.V., Plourde, S.M., Rand, T.G., Soderhall, K. and Gutell, R.R. (1996) A novel clade of protistan parasites near the animal-fungal divergence. Proc. Natl. Acad. Sci. USA, 93, 11907-11912.
Ralph, S.A., Van Dooren, G.G., Waller, R.F., Crawford, M.J., Fraunholz, M.J., Foth, B.J., Tonkin, C.J., Roos, D.S. and McFadden, G.I. (2004) Tropical infectious diseases: Metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat. Rev. Microbiol., 2, 203-216.
Reichert, A.S. and Neupert, W. (2004) Mitochondriomics or what makes us breathe. Trends Genet., 20, 555-562.
Reyes-Prieto, A., Weber, A.P. and Bhattacharya, D. (2007) The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet., 41, 147-168.
Rice, D.W. and Palmer, J.D. (2006) An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol., 4, 31.
Richards, T.A. and Cavalier-Smith, T. (2005) Myosin domain evolution and the primary divergence of eukaryotes. Nature, 436, 1113-1118.
Rodriguez-Ezpeleta, N., Brinkmann, H., Burey, S.C., Roure, B., Burger, G., Loffelhardt, W., Bohnert, H.J., Philippe, H. and Lang, B.F. (2005) Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol, 15, 1325-1330.
Rodriguez-Ezpeleta, N., Brinkmann, H., Burger, G., Roger, A.J., Gray, M.W., Philippe, H. and Lang, B.F. (2007) Toward resolving the eukaryotic tree: the phylogenetic positions of jakobids and cercozoans. Curr. Biol., 17, 1420-1425.
Roger, A.J. (1999) Reconstructing early events in eukaryotic evolution. Am. Nat., 154, S146-S163.
Rogers, M.B., Gilson, P.R., Su, V., McFadden, G.I. and Keeling, P.J. (2007) The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol. Biol. Evol., 24, 54-62.
Rohdich, F., Kis, K., Bacher, A. and Eisenreich, W. (2001) The non-mevalonate pathway of isoprenoids: genes, enzymes and intermediates. Curr. Opin. Chem. Biol., 5, 535-540.
Ruiz-Trillo, I., Lane, C.E., Archibald, J.M. and Roger, A.J. (2006) Insights into the evolutionary origin and genome architecture of the unicellular opisthokonts Capsaspora owczarzaki and Sphaeroforma arctica. J. Eukaryot. Microbiol., 53, 379-384.
Rumpho, M.E., Worful, J.M., Lee, J., Kannan, K., Tyler, M.S., Bhattacharya, D., Moustafa, A. and Manhart, J.R. (2008) Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica. Proc. Natl. Acad. Sci. USA, 105, 17867-17871.
Simpson, A.G. (2003) Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota). Int. J. Syst. Evol. Microbiol., 53, 1759-1777.
Simpson, A.G., Inagaki, Y. and Roger, A.J. (2006) Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of "primitive" eukaryotes. Mol. Biol. Evol., 23, 615-625.
Simpson, A.G. and Patterson, D.J. (2001) On core jakobids and excavate taxa: the ultrastructure of Jakoba incarcerata. J. Eukaryot. Microbiol., 48, 480-492.
Simpson, A.G. and Roger, A.J. (2002) Eukaryotic evolution: getting to the root of the problem. Curr Biol, 12, R691-693.
Simpson, A.G., Roger, A.J., Silberman, J.D., Leipe, D.D., Edgcomb, V.P., Jermiin, L.S., Patterson, D.J. and Sogin, M.L. (2002) Evolutionary history of "early-diverging" eukaryotes: the excavate taxon Carpediemonas is a close relative of Giardia. Mol. Biol. Evol., 19, 1782-1791.
Simpson, A.G.B. and Patterson, D.J. (1999) The ultrastructure of Carpediemonas membranifera (Eukaryota) with reference to the "Excavate hypothesis". Eur. J. Protistol., 35, 353-370.
Sogin, M.L. (1989) Evolution of eukaryotic microorganisms and their small subunit ribosomal RNAs. Amer. Zool., 29, 487-499.
Sogin, M.L., Elwood, H.J. and Gunderson, J.H. (1986) Evolutionary diversity of eukaryotic small-subunit rRNA genes. Proc. Natl. Acad. Sci. USA, 83, 1383-1387.
Stanier, R.Y. (1970) Some aspects of the biology of cells and their possible evolutionary significance. Symp. Soc. Gen. Mircrobiol., 20, 1-38.
Stechmann, A. and Cavalier-Smith, T. (2002) Rooting the eukaryote tree by using a derived gene fusion. Science, 297, 89-91.
Stechmann, A. and Cavalier-Smith, T. (2003) The root of the eukaryote tree pinpointed. Curr. Biol., 13, R665-666.
Steenkamp, E.T., Wright, J. and Baldauf, S.L. (2006) The protistan origins of animals and fungi. Mol. Biol. Evol., 23, 93-106.
Taylor, F.J. (1978) Problems in the development of an explicit hypothetical phylogeny of the lower eukaryotes. Biosystems, 10, 67-89.
Tovar, J., Fischer, A. and Clark, C.G. (1999) The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol. Microbiol., 32, 1013-1021.
Turner, S. (1997) Molecular systematics of oxygenic photosynthetic bacteria. Pl. Syst. Evol. [Suppl.], 11, 13-52.
Turner, S., Pryer, K.M., Miao, V.P. and Palmer, J.D. (1999) Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol., 46, 327-338.
van der Giezen, M. and Tovar, J. (2005) Degenerate mitochondria. EMBO Rep., 6, 525-530.
van der Giezen, M., Tovar, J. and Clark, C.G. (2005) Mitochondrion-derived organelles in protists and fungi. Int. Rev. Cytol., 244, 175-225.
Wainright, P.O., Hinkle, G., Sogin, M.L. and Stickel, S.K. (1993) Monophyletic origins of the metazoa: an evolutionary link with fungi. Science, 260, 340-342.
Williams, B.A.P. and Keeling, P.J. (2003) Cryptic organelles in parasitic protists and fungi. Adv. Parasitol., 54, 9-67.
Wilson, R.J. (2002) Progress with parasite plastids. J. Mol. Biol., 319, 257-274.
Woese, C.R. (1987) Bacterial evolution. Microbiol. Rev., 51, 221-271.
Woese, C.R. and Fox, G.E. (1977) Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA, 74, 5088-5090.
Woese, C.R., Kandler, O. and Wheelis, M.L. (1990) Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA, 87, 4576-4579.
Yoon, H.S., Hackett, J.D., Pinto, G. and Bhattacharya, D. (2002) A single, ancient origin of the plastid in the Chromista. Proc. Natl. Acad. Sci. USA, 99, 15507-15512.
Information on the Internet
- Eu-Tree. Assembling the Tree of Eukaryotic Diversity.
- 'Tree Of Life' Has Lost A Branch, According To Largest Genetic Comparison Of Higher Life Forms Ever. Science Daily.
- Protsville. Protist Research Laboratory, University of Sydney, Australia.
- Protist Information Server. Japan Science and Technology Corporation.
- Eukaryota: Systematics. Museum of Paleontology, University of California, Berkeley, USA.
- Malaria, Algae, Amoeba and You: Unravelling Eukaryotic Relationships. Joel B. Dacks. ActionBioScience.org
- Exploring Early Eukaryotic Evolution: Diversity and Relationships Among Novel Deep-Branching Lineages . Virginia Edgcomb, Andrew Roger, Alastair G.B. Simpson, Jeffrey Silberman and Mitchell Sogin, Marine Biological Laboratory, Woods Hole, USA.
- Microbial Life - Educational Resources. Teaching and learning about the diversity, ecology and evolution of the microbial world; discover the connections between microbial life, the history of the earth and our dependence on micro-organisms.
- Eukaryotes in extreme environments. Dave Roberts, the Natural History Museum, London, UK.
- The Homeobox Page. Thomas R. Bürglin's page about the homeobox genes which play important roles in the development of multicellular organisms.
- Protist Image Data. Molecular Evolution and Organelle Genomics program at the University of Montreal, Canada.
Title Illustrations

| Location | along trail at night, La Selva Biological Station, lowland rain forest of Costa Rica |
|---|---|
| Specimen Condition | Live Specimen |
| Copyright |
© Greg and Marybeth Dimijian

|
| Specimen Condition | Scanning Electron Micrograph |
|---|---|
| Image Use |
 This media file is licensed under the Creative Commons Attribution-NonCommercial License - Version 3.0. This media file is licensed under the Creative Commons Attribution-NonCommercial License - Version 3.0.
|
| Copyright |
© Brian S. Leander

|
| Scientific Name | Stephanopogon minuta (Heterolobosea) |
|---|---|
| Location | Vancouver, British Columbia |
| Specimen Condition | Scanning Electron Micrograph |
| Copyright | © Naoji Yubuki and Brian Leander |
| Scientific Name | Phacus brachykentron |
|---|---|
| Specimen Condition | Scanning electron micrograph |
| Copyright |
© Brian S. Leander

|
| Scientific Name | Heliconia sp. |
|---|---|
| Location | cultivated, North Carolina Zoo |
| Specimen Condition | Live Specimen |
| Source | http://www.flickr.com/photos/ucumari/533255173/ |
| Source Collection | Flickr |
| Image Use |
 This media file is licensed under the Creative Commons Attribution-NonCommercial-NoDerivs License - Version 2.0. This media file is licensed under the Creative Commons Attribution-NonCommercial-NoDerivs License - Version 2.0.
|
| Copyright | © 2007 Valerie |
About This Page
This page is being developed as part of the Tree of Life Web Project Protist Diversity Workshop, co-sponsored by the Canadian Institute for Advanced Research (CIFAR) program in Integrated Microbial Biodiversity and the Tula Foundation.
Patrick Keeling

The University of British Columbia, Vancouver, British Columbia, Canada
Brian S. Leander

The University of British Columbia, Vancouver, British Columbia, Canada
Correspondence regarding this page should be directed to Patrick Keeling at
pkeeling@interchange.ubc.ca
and Brian S. Leander at
bleander@interchange.ubc.ca
Page copyright © 2009 Patrick Keeling and Brian S. Leander
 Page: Tree of Life
Eukaryotes. Eukaryota, Organisms with nucleated cells.
Authored by
Patrick Keeling and Brian S. Leander.
The TEXT of this page is licensed under the
Creative Commons Attribution-NonCommercial License - Version 3.0. Note that images and other media
featured on this page are each governed by their own license, and they may or may not be available
for reuse. Click on an image or a media link to access the media data window, which provides the
relevant licensing information. For the general terms and conditions of ToL material reuse and
redistribution, please see the Tree of Life Copyright
Policies.
Page: Tree of Life
Eukaryotes. Eukaryota, Organisms with nucleated cells.
Authored by
Patrick Keeling and Brian S. Leander.
The TEXT of this page is licensed under the
Creative Commons Attribution-NonCommercial License - Version 3.0. Note that images and other media
featured on this page are each governed by their own license, and they may or may not be available
for reuse. Click on an image or a media link to access the media data window, which provides the
relevant licensing information. For the general terms and conditions of ToL material reuse and
redistribution, please see the Tree of Life Copyright
Policies.
- First online 08 September 2000
- Content changed 28 October 2009
Citing this page:
Keeling, Patrick and Brian S. Leander. 2009. Eukaryotes. Eukaryota, Organisms with nucleated cells. Version 28 October 2009. http://tolweb.org/Eukaryotes/3/2009.10.28 in The Tree of Life Web Project, http://tolweb.org/


















 Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site
Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site