IRLC (Inverted Repeat-lacking clade)
Martin F. Wojciechowski.png?x=-126624561)

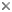
This tree diagram shows the relationships between several groups of organisms.
The root of the current tree connects the organisms featured in this tree to their containing group and the rest of the Tree of Life. The basal branching point in the tree represents the ancestor of the other groups in the tree. This ancestor diversified over time into several descendent subgroups, which are represented as internal nodes and terminal taxa to the right.

You can click on the root to travel down the Tree of Life all the way to the root of all Life, and you can click on the names of descendent subgroups to travel up the Tree of Life all the way to individual species.
For more information on ToL tree formatting, please see Interpreting the Tree or Classification. To learn more about phylogenetic trees, please visit our Phylogenetic Biology pages.
close boxIntroduction
The Inverted Repeat-lacking clade or IRLC (Wojciechowski et al., 2000), so-called because it is uniquely marked by the loss of one copy of the large inverted repeat (approx. 25 kilobase) in the chloroplast genome (see below, under Characteristics), includes most members of Polhill's (1981) "temperate herbaceous group" of papilionoids. This group comprises all members of tribes Cicereae, Hedysareae, Trifolieae, and Fabeae (also known as Vicieae), as well as at least three genera, Afgekia Craib., Callerya Endl., and Wisteria Nutt., currently treated in tribe Millettieae (Schrire, 2005), nested within the paraphyletic tribe Galegeae (Polhill, 1994).
Polhill's "temperate herbaceous group" of tribes had been distinguished from other predominantly temperate tribes such as Thermopsideae (e.g., genera such as Baptisia, Thermopsis) by the accumulation of the non-protein amino acid canavanine, rather than alkaloids, in seeds. This group contains many of the familiar temperate and agriculturally important legumes such as alfalfa, clovers, lentils, chickpea, garden pea, vetches, as well as locoweeds, and ornamental "desert peas" and wisterias. The IRLC also includes a large number of model species such as Medicago truncatula (barrel medic), Medicago sativa (alfalfa), Pisum sativum (garden pea), Vicia faba (fava bean), and Trifolium repens (white clover), used for studies of symbiotic nitrogen fixation, root nodule development, legume development, genetics and genomics, and bacterial-plant coevolution.
Characteristics
The IRLC is dominated by often large, temperate genera such as Astragalus L., Hedysarum L., Medicago L., Oxytropis DC., Swainsona Salisb., and Trifolium L., which share a number of morphological characters including a predominantly herbaceous habit (annual and perennial), epulvinate compound leaves, stipules adnate to the petiole, base chromosome numbers of n = 7 or n = 8, and centers of greatest species diversity in Eurasia and North America (Polhill and Raven, 1981; Polhill, 1994), in addition to the loss of one copy of the inverted repeat in the chloroplast genome. This structural mutation in legumes (Palmer et al., 1987; Lavin et al., 1990) is particularly remarkable since the inverted repeat, which encodes a duplicate set of ribosomal RNA genes (structural RNA molecules used to make ribosomes which are part of the cell machinery for protein synthesis), is an evolutionary conserved feature of green algal and land plant chloroplast genomes (Palmer et al., 1988), and is known to be absent otherwise only from certain conifers (Strauss et al., 1988) and a few, specific genera in the angiosperm families Geraniaceae and Orobanchaceae (Downie and Palmer, 1992).
Discussion of Phylogenetic Relationships
The IRLC was the first clade within legumes essentially distinguished on the basis of a molecular synapomorphy, the loss of one copy of the 25-kb IR in the chloroplast genome. The monophyly of the IRLC has been consistently and strongly supported (e.g., 100% bootstrap proportions) in essentially all studies based on cladistic analyses of molecular data, including chloroplast DNA restriction fragment length polymorphisms (Lavin et al., 1990; Liston, 1995), nuclear rDNA ITS sequences (Sanderson and Wojciechowski, 1996) and chloroplast gene/intron sequences (Doyle et al., 1997; Käss and Wink, 1997; Hu et al., 2000; Kajita et al., 2001; Wojciechowski et al., 2004).
Within the IRLC, the so-called "IRLC millettioids", genera Afgekia, Callerya, and Wisteria (and possibly Endosamara) along with Glycyrrhiza of Galegeae form a paraphyletic grade at the base of the IRLC. Relationships among these lineages are currently not well-resolved or supported, a consequence most likely due to the lack of adequate sampling. Three well-supported subclades, the "Hedysaroid" clade, Galegeae sens. lat., and the "Vicioid" clade, comprise the remainder of the IRLC. The tribe Hedysareae (Polhill, 1994), recently expanded by Lock (2005) to include the genera Calophaca, Caragana, Halimodendron, and Alhagi formerly treated in Galegeae (Polhill, 1994) in addition to the large genera Hedysarum and Onobrychis, comprises the Hedysaroid clade. However, sampling within the Hedysaroid clade has been limited and relationships both within and among the 12 genera (and 400-450 species) remain poorly understood.
The Galegeae sens. lat. subclade of the IRLC consists of the majority of genera formerly treated in tribe Galegeae (sensu Polhill, 1994; Lock and Schrire, 2005) and include Astragalus, Chesneya, Oxytropis, Sutherlandia, Swainsona, Colutea, and Carmichaelia. With the exception of Astragalus and Oxytropis, all of these genera are distributed exclusively in Eurasia, Africa, or Australasia. Nested within this group is the well-supported "Astragalean" clade which includes the genus Astragalus, the largest genus of vascular plants with an estimated 2,500 species (plus a number of segregates), Oxytropis, and subtribe Colutinae (Wojciechowsk et al., 1999, 2000).
The Vicioid subclade of the IRLC includes many of the agriculturally important genera and model species such as Cicer, Medicago, Pisum, Trifolium, and Vicia. Within this subclade, the genus Parochetus (Trifolieae) is consistently resolved as the sister group to all other vicioid taxa; genus Galega forms the sister group to the monogeneric tribe Cicereae (Cicer), and together these taxa form the sister group to a clade that includes the tribes Trifolieae and Fabeae (formely known as Vicieae) (Steele and Wojciechowski, 2003; Wojciechowski et al., 2004). Results based on analyses of the matK gene (Steele and Wojciechowski, 2003) suggest the genus Trifolium ("clovers") is the sister group to the Fabeae rather than to remaining members of the tribe Trifolieae, but this position is still weakly supported and the subject of on-going investigation (e.g., see recent phylogeny of Trifolium; Ellison et al., 2006).
References
Downie, S. R., and J. D. Palmer. 1992. Use of chloroplast DNA rearrangements in reconstructing plant phylogeny. Pages 14-35 in Molecular Systematics of Plants (P. S. Soltis, D. E. Soltis, and J. J. Doyle, eds.). Chapman and Hall, New York, NY.
Doyle, J.J., J.L. Doyle, J.A. Ballenger, E.E. Dickson, T. Kajita, and H. Ohashi. 1997. A phylogeny of the chloroplast gene rbcL in the Leguminosae: taxonomic correlations and insights into the evolution of nodulation. American J. Botany 84: 541-554.
Ellison, N. W., A. Liston, J. J. Steiner, W. M. Williams, and N. L. Taylor. 2006. Molecular phylogenetics of the clover genus Trifolium (Leguminosae). Molecular Phylogenetics and Evolution 39: 688-705.
Hu, J.-M., M. Lavin, M. F. Wojciechowski, and M.J. Sanderson. 2000. Phylogenetic systematics of the tribe Millettieae (Leguminosae) based on matK sequences, and implications for evolutionary patterns in Papilionoideae. American J. Botany 87: 418-430.
Kajita, T., H. Ohashi, Y. Tateishi, C. D. Bailey, and J. J. Doyle. 2001. rbcL and legume phylogeny, with particular reference to Phaseoleae, Millettieae, and allies. Systematic Botany 26: 515-536.
Käss, E., and M. Wink. 1997. Phylogenetic relationships in the Papilionoideae (Family Leguminosae) based on nucleotide sequences of cpDNA (rbcL) and ncDNA (ITS1 and 2). Molecular Phylogenetics and Evolution 8:65-88.
Lavin, M., J. J. Doyle, and J. D. Palmer. 1990. Evolutionary significance of the loss of the chloroplast--DNA inverted repeat in the Leguminosae subfamily Papilionoideae. Evolution 44: 390-402.
Liston, A. 1995. Use of the polymerase chain reaction to survey for the loss of the inverted repeat in the legume chloroplast genome. Pages 31-40 in Advances in legume systematics, part 7, Phylogeny (M. D. Crisp and J. J. Doyle, eds.). Royal Botanic Gardens, Kew, UK.
Lock, J. M. 2005. Hedysareae. Pages 489-495 in Legumes of the world (Lewis et al., eds.). Royal Botanic Gardens, Kew, UK.
Lock, J. M., and B. D. Schrire. 2005. Galegeae. Pages 475-487 in Legumes of the world (Lewis et al., eds.). Royal Botanic Gardens, Kew, UK.
Palmer, J. D., B. Osorio, J. Aldrich, and W. F. Thompson. 1987. Chloroplast DNA evolution among legumes: loss of a large inverted repeat occurred prior to other sequence rearrangements. Current Genetics 11: 275-286.
Palmer, J. D., R. K. Jansen, H. J. Michaels, M. W. Chase, and J. R. Manhart. 1988. Chloroplast DNA variation and plant phylogeny. Annals of the Missouri Botanical Garden 75: 1180-1206.
Polhill, R. M. 1981. Papilionoideae. Pages 191-208 in Advances in Legume Systematics, part 1 (R. M. Polhill and P. Raven, eds.). Royal Botanic Gardens, Kew, UK.
Polhill, R. M. 1994. Classification of the Leguminosae. Pages xxxv–xlviii in Phytochemical Dictionary of the Leguminosae (F. A. Bisby, J. Buckingham, and J. B. Harborne, eds.). Chapman and Hall, New York, NY.
Polhill, R. M., and P. H. Raven (eds.). 1981. Advances in legume systematics, parts 1 and 2. Royal Botanic Gardens, Kew, UK.
Schrire, B. D. 2005. Millettieae. Pages 367-387 in Legumes of the world (Lewis et al., eds.). Royal Botanic Gardens, Kew, UK.
Sanderson, M. J., and M. F. Wojciechowski. 1996. Diversification rates in a temperate legume clade: are there "so many species" of Astragalus (Fabaceae)? American J. Botany 83: 1488-1502.
Steele, K. P., and M. F. Wojciechowski. 2003. Phylogenetic analyses of tribes Trifolieae and Vicieae based on sequences of the plastid gene matK (Papilionoideae: Leguminosae). Pages 355-370 in Advances in Legume Systematics, part 10, higher level systematics (B. B. Klitgaard and A. Bruneau, eds.). Royal Botanic Gardens, Kew, UK.
Strauss, S. H., J. D. Palmer, G. T. Howe and A. H. Doerksen. 1988. Chloroplast genomes of two conifers lack a large inverted repeat and are extensively rearranged. Proceedings of the National Academy of Sciences USA 85: 3898-3902.
Wojciechowski, M. F., M. Lavin, and M. J. Sanderson. 2004. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. American J. Botany 91: 1846-1862.
Wojciechowski, M. F., M. J. Sanderson, and J.-M. Hu. 1999. Evidence on the monophyly of Astragalus (Fabaceae) and its major subgroups based on nuclear ribosomal DNA ITS and chloroplast DNA trnL intron data. Systematic Botany 24: 409–437.
Wojciechowski, M. F., M. J. Sanderson, K. P. Steele, and A. Liston. 2000. Molecular phylogeny of the “temperate herbaceous tribes” of papilionoid legumes: a supertree approach. Pages 277-298 in Advances in Legume Systematics, part 9 (P. S. Herendeen and A. Bruneau, eds.). Royal Botanic Gardens, Kew, UK.
Title Illustrations

| Scientific Name | Medicago sativa L. |
|---|---|
| Specimen Condition | Live Specimen |
| Copyright | © 2006 International Legume Research Institute |
| Scientific Name | Astragalus purshii Dougl. ex G.Don |
|---|---|
| Location | California, USA |
| Specimen Condition | Live Specimen |
| Copyright | © 2006 Jay Sullivan |
| Scientific Name | Pisum sativum L. |
|---|---|
| Location | Island of Mallorca, Spain |
| Specimen Condition | Live Specimen |
| Copyright | © 2006 Jardin Mundani |
| Scientific Name | Wisteria sinensis (Sims) Sweet |
|---|---|
| Specimen Condition | Live Specimen |
| Copyright | © 2006 Annette Höggemeier |
About This Page
Martin F. Wojciechowski

Arizona State University, Tempe, Arizona, USA
Correspondence regarding this page should be directed to Martin F. Wojciechowski at
mfwojciechowski@asu.edu
Page copyright © 2006 Martin F. Wojciechowski
All Rights Reserved.
- First online 11 July 2006
- Content changed 11 July 2006
Citing this page:
Wojciechowski, Martin F. 2006. IRLC (Inverted Repeat-lacking clade). Version 11 July 2006 (under construction). http://tolweb.org/IRLC_%28Inverted_Repeat-lacking_clade%29/60358/2006.07.11 in The Tree of Life Web Project, http://tolweb.org/










 Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site
Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site