Heterolobosea
Johan F. De Jonckheere

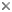
This tree diagram shows the relationships between several groups of organisms.
The root of the current tree connects the organisms featured in this tree to their containing group and the rest of the Tree of Life. The basal branching point in the tree represents the ancestor of the other groups in the tree. This ancestor diversified over time into several descendent subgroups, which are represented as internal nodes and terminal taxa to the right.

You can click on the root to travel down the Tree of Life all the way to the root of all Life, and you can click on the names of descendent subgroups to travel up the Tree of Life all the way to individual species.
For more information on ToL tree formatting, please see Interpreting the Tree or Classification. To learn more about phylogenetic trees, please visit our Phylogenetic Biology pages.
close boxThis tree is based on Nikolaev et al. 2004, Park et al. 2007 and Yubuki and Leander, in press.
F - flagellate, A - amoeba, AF - amoeboflagellate.
ATCC: American Type Culture Collection
Introduction
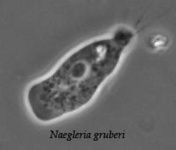
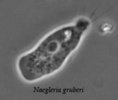
The class Heterolobosea (Page and Blanton 1985) has been established for amoebae (A) that move with eruptive bulges, have intranuclear orthomitosis, show transformation to a temporary flagellate stage (F) (Fig. 1) in some genera, have mitochondrial cristae flattened, often disc-like (Fig. 2), and a Golgi system not organized as stacks of flattened cisternae.

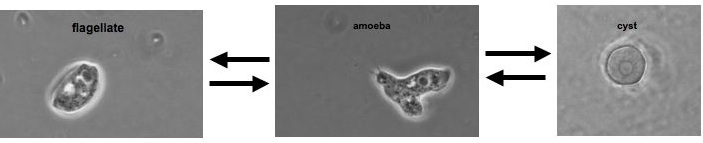
Fig. 1. Life cycle of Naegleria gruberi. © Johan F. De Jonckheere

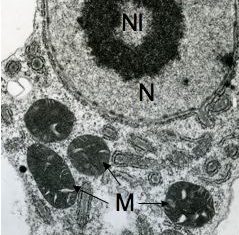
Fig. 2. EM of Naegleria lovaniensis. M: mitochondria, N: nucleus, Nl: nucleolus. © Johan F. De Jonckheere
The Heterolobosea are traditionally subdivided into two orders, the Schizopyrenida and the Acrasida, distinguished from each other by the formation of fruiting bodies in the latter (Page and Blanton 1985). The order Schizopyrenida is divided into two families, the Vahlkampfiidae, which is clearly polyphyletic (Nikolaev et al. 2004, Park et al. 2007, Yubuki and Leander, in press), and the Gruberellidae.
For a long time most research on Heterolobosea remained based on morphology. Traditionally (Page 1988) Vahlkampfiidae with no flagellate stage were classified in the genus Vahlkampfia, while those with a flagellate stage were either Naegleria (including Adelphamoeaba (De Jonckheere 2002)) with biflagellate stage, or Willaertia (probably corresponding to Tetramastigamoeba) and Tetramitus (including Paratetramitus (De Jonckheere and Brown 2005)) with quadriflagellated stage.
With the recognition of a human pathogen, Naegleria fowleri, in the family Vahlkampfiidae many new techniques were used to investigate the genus Naegleria. Using DNA sequencing, many cryptic species (De Jonckheere 2002) and even cryptic genera were discovered in the Vahlkampfiidae (Brown and De Jonckheere 1999). It became clear that not all species in the amoeboflagellate (AF) genus Naegleria could transform to flagellates with two flagella, but that there were strains or species that could not transform or did so with difficulty under normal conditions. Also Naegleria spp. were identified in which the flagellates could divide (De Jonckheere 2002). Dividing flagellates with four flagella were also found in the newly established Willaertia magna (Robinson et al. 1989), sharing this characteristic with the genus Tetramitus. However, in the genus Tetramitus species were subsequently described which also did not form flagellates (De Jonckheere and Brown, 2005) or did so with difficulty (Robinson et al. 2007).
Not much progress has been made in the Gruberellidae, and there is only molecular support for the inclusion of Stachyamoeba in the Heterolobosea, while there are no data for Gruberella. Although there is molecular support for the inclusion of Acrasis rosea in the Heterolobosea, the Acrasida remained poorly studied during the last 20 years.
Recently it has been established with molecular methods that Stephanopogon, which remained for decades an enigmatic organism and was placed in the Pseudociliata, phylogenetically belongs to the Heterolobosea (Yubuki and Leander 2008) and its closest relative is the marine flagellate Percolomonas cosmopolitus, which was also found previously to belong to the Heterolobosea (Nikolaev et al. 2004).
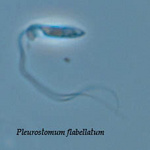
Four extremophiles have been reported in the Heterolobosea: Tetramitus thermacidophilus (Baumgartner et al. in press), Pleurostomum flabellatum (Park et al. 2007), and Macropharyngomonas halophila (AF011465, ATCC 50621) and Plaesiobystra hypersalinica (AF011459, ATCC 50619), but the latter two are not formally described.
Characteristics
Vahlkampfiidae
In vahlkampfiids, the nucleolus forms polar masses in mitosis. Vahlkampfiids are normally uninucleate, and in some genera transformation from amoeba to flagellate occurs. Many genera and species are known.
In the genus Naegleria alone 47 different species have been distinguished mostly based on differences in rDNA sequences. The genus is subdivided into 8 clusters, of which two contain species which are only found in cold environments (De Jonckheere 2006b). The genus Naegleria is notorious because the species N. fowleri causes primary amoebic meningoencephalitis (PAM), almost invariably leading to death. Cases of PAM are detected worldwide and are mostly related to swimming in warm water, whereby the pathogen enters the brain via the nose. The pathogen N. fowleri grows at a temperature up to 45°C and shares this characteristic with the closely related N. lovaniensis. Therefore, these two species can be found in warm waters where other species cannot grow. Two other species, N. australiensis and N. italica, were found to be pathogenic in experimental animals, but no human cases are known yet (De Jonckheere 2002). Most Naegleria spp. transform into flagellates with two flagella but nonflagellating species are known. Most species have flagellates which do not divide but in a few species division in the flagellate stage is observed. Almost all species form cysts with pores. There is some variation in cyst morphology (Fig. 3).

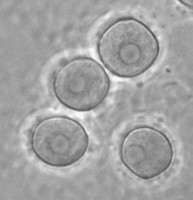
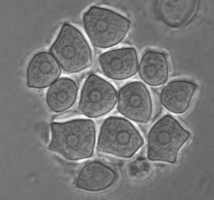
Fig. 3. Cysts of Naegleria gruberi and Naegleria angularis. © Johan F. De Jonckheere
Willaertia magna was originally regarded as belonging to the genus Naegleria based essentially on cyst morphology. Molecular phylogeny showed, however, that it is a different genus, but it is the most closely related to the Naegleria genus. The cysts of Willaertia magna have pores (Fig. 4) as in Naegleria spp. and the flagellates with four flagella divide successively resulting in progressive reduction in cell volume (Robinson et al. 1989). This successive division has not been observed in Naegleria spp. in which dividing flagellates are detected.

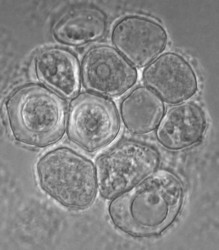
Fig. 4. Cysts of Willaertia magna.© Johan F. De Jonckheere
All Vahlkampfiidae which did not transform into flagellates were originally placed in the genus Vahlkampfia (Page 1988). Upon the subsequent observation of flagellate formation in V. jugosa, this species was moved to a new genus, Paratetramitus. Molecular analysis of the rDNA showed, however, that also other Vahlkampfia spp. did not belong to this genus (Brown and De Jonckheere 1999). Some were moved to newly established genera, i.e. Paravahlkampfia and Neovahlkampfia, while others turned out to belong to the genus Tetramitus, although no transformation to the flagellate stage was observed in these species. As such the genera Didascalus, Singhamoeba and Learamoeba turned out to be junior synonyms of Tetramitus (De Jonckheere and Brown 2005). Also Paratetramitus jugosus had to be moved to the genus Tetramitus. As such the genus Tetramitus contains species that can or cannot transform to the flagellate stage, as is the case in the genus Naegleria. Although all belong to the genus Tetramitus the cyst morphology varies much more between Tetramitus spp. (Fig. 5) than in the genus Naegleria. The cyst can have a smooth wall or wrinkled outer wall, both with no pores such as in T. wacamawensis, T. ovis and T. jugosus, one pore such as in T. horticolus, or several pores such as in T. angularis. This high variation in cyst morphology lead some investigators to erect different genera for those isolates although molecular data contradict this.

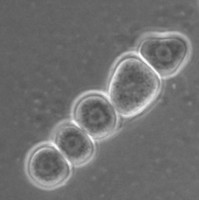
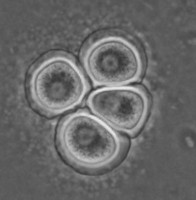
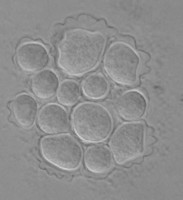
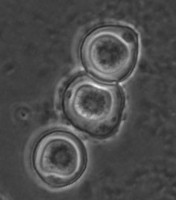
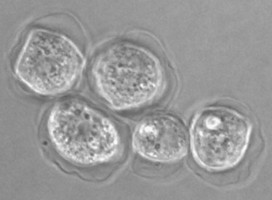
Fig. 5. Cysts of Tetramitus spp. Top: Tetramitus wacamawensis, Tetramitus ovis, and Tetramitus jugosus. Bottom: Tetramitus horticolus and Tetramitus angularis. © Johan F. De Jonckheere.
In the genus Tetramitus an extremophile has been detected, T. thermacidophilus which grows in a pH range of 1.2 – 5 and a temperature up to 54°C (Baumgartner et al. submitted).
Later isolates of the vahlkampfiids turned out to belong mostly to the genus Tetramitus (at present 15 species are recognized in the genus Tetramitus), although one was a new species of Paravahlkampfia (Brown and De Jonckheere, 2004) and a few new isolates belong phylogenetically to the genus Vahlkampfia, i. e. V. signyensis (Garstecki et al. 2005), V. ciguana and V. orchilla (De Jonckheere 2006a) and their cysts all look very similar (Fig. 6).

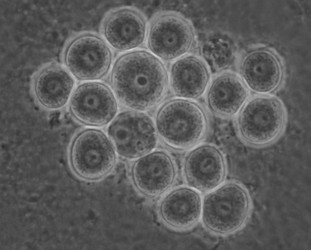
Fig. 6. Cysts of Vahlkampfia signyensis.© Johan F. De Jonckheere
A recent marine amoeba isolate, EV10A (ATCC N° PRA-14), constitutes a new genus which is most closely related to Willaertia (Baumgartner et al. in preparation).
Neovahlkampfia damariscottae, Heteramoeba clara and Pernina chaumonti are the only marine Vahlkampfiidae, although for the genus Pernina there is no molecular evidence for its inclusion in the Vahlkampfiidae. As the type strain of Pernina (El Kadiri et al. 1992) is lost, no such evidence can be obtained. The genus Heteramoeba clusters with the halophilic Plaesiobystra, not formally described, but Neovahlkampfia seems not to be closely related with other marine or halophilic genera. The marine amoeba Neovahlkampfia damariscottae clusters in SSU rDNA phylogeny (AY965861) with an amoeba (strain And 9) isolated from soil (Lara et al. 2007).
Pleurostomum flabellatum is a flagellate (Fig. 7) of which no amoebic form is reported. It is an extreme halophile but is closely related to Naegleria and Willaertia (Park et al. 2007), which are genera only present in freshwater. However, discoid mitochondrial cristae, typical of the Heterolobosea, were not observed in Pleurostomum.

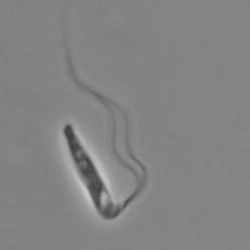
Fig. 7. Pleurostomum flabellatum. © Alastair Simpson
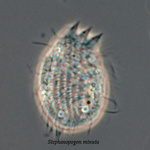
Another flagellate, which does not have an amoeba stage, Percolomonas cosmopolitus, was also shown to belong to the Heterolobosea (Nikolaev et al. 2004) and its closest relative turned out to be Stephanopogon (Yubuki and Leander 2008). Stephanopogon spp. (Fig. 8) differ from the other flagellates in the Heterolobosea by their huge number of flagella (112 to 144), the reason why they were previously classified as Pseudociliata. In Stephanopogon, in which six species have been described, the lack of Golgi bodies and the presence of discoidal mitochondrial cristae indicate that this genus belongs indeed to the Heterolobosea and the SSU rDNA analysis supports this.
In Percolomonas cosmopolitus the SSU rDNA analysis shows a high divergence between the two strains analyzed (Nikolaev et al. 2004) which might indicate the presence of cryptic species.

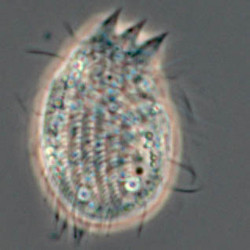
Fig. 8. Stephanopogon minuta. © Naoji Yubuki & Brian S. Leander
Monopylocystis visvesvarai and Sawyeria marylandensis, two amitochondrial, anaerobic or microaerophilic amoebae, with no flagellate stage, are related to the microaerophilic amoeboflagellate Psalteriomonas lanterna, within the Heterolobosea. Although some would like to include these in a separate grouping, the Lyromonadea, Psalteriomonas is characterized by a unique insertion (helix 17_1) in the SSU rDNA secondary structure (Nikolaev et al. 2004), which is characteristic for all the Heterolobosea, while structural data of the flagellar apparatus confirm the relationship (Brugerolle and Simpson 2004).
Pseudomastigamoeba longifilum is an undetermined Heterolobosean because the source organism (ATCC 50575) of which the SSU rDNA sequence was determined could not be confirmed and the sequence (N° AF11462) was removed from GenBank (Nikolaev et al. 2004).
Macropharyngomonas halophila, also not formally described, does not have the helix 17_1 typical of the Heterolobosea, but the basal position in the phylogenetic tree suggests this helix appeared after the divergence of M. halophila (Nikolaev et al. 2004).
Gruberellidae
The Gruberellidae are distinguished from the Vahlkampfiidae by the disintegration of the nucleolus during mitosis. Two genera and species are known.
The genus and species Stachyamoeba lipophora was isolated from soil (page 1988). No molecular analysis of this isolate was performed and the type strain held by CCAP (Culture Collection of Algae and Protozoa) was lost, precluding any further phylogenetic investigation. The molecular analysis (AF011461) of a less well described Stachyamoeba sp. from seawater showed that this ATCC strain (Fig. 9) belongs to the Heterolobosea. It is uncertain, however, whether this ATCC strain is related to the type strain of Stachyamoeba because of the very different origin, soil versus seawater. Also we recently detected flagellate (Fig. 9c) transformation in this ATCC Stachyamoeba strain (Murase and De Jonckheere, in preparation), which was not reported for the type strain of Stachyamoeba. Recently, two isolates, an soil amoeba strain 6-5F (Murase and De Jonckheere, in preparation) and a marine amoeboflagellate strain SAN2 (ATCC N° PRA-15) (Baumgartner et al, in preparation) have shown to be closely related to the ATCC strain of Stachyamoeba.
Another genus and species in the Gruberellidae, Gruberella flavescens, is only known from the marine environment, is usually multinucleate, does not form flagellates and no cyst were detected (Page 1983). There is no molecular confirmation of the phylogenetic relationship with Stachyamoeba.

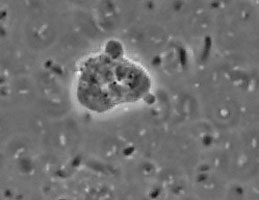
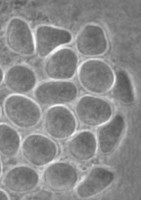
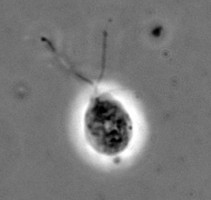
Fig. 9. a. Amoeba and b. cysts of Stachyamoeba sp. ATCC 50324. © Johan F. De Jonckheere. c. Flagellate of Stachyamoeba sp. ATCC 50324. © Jun Murase
Acrasida
As in the Gruberellidae the nucleolus disintegrates during mitosis, the amoebae of some species can transform into flagellates but in the Acrasida the amoeba also form fruiting bodies. Phylogenetic analysis of the SSU rDNA (AF011458) does indicate that Acrasis rosea belongs to the Heterolobosea and it carries that unique insertion (helix 17_1) in the SSU rDNA secondary structure typical for the Heterolobosea (Nikolaev et al. 2004). A recent amoeba isolate from soil (strain And 12) clusters in SSU rDNA phylogeny (AY965862) with Acrasis rosea (Lara et al. 2007).
Of all other genera in the Acrasida, such as Guttulinopsis, Pocheina and Rosculus (Page 1988), there is no molecular information on their phylogeny.
rDNA and Group I Introns
In different species of Naegleria and Tetramitus the rDNA genes are on a circular plasmid, which was estimated to be present in 4,000 copies (Clark and Cross 1988). It is not known whether this rDNA configuration is also present in the Heterolobosea other than the Vahlkampfiidae. In several species of Naegleria, a group I intron is present in the SSU rDNA, and in a few species also in the LSU rDNA (De Jonckheere 2002, 2004). Group I introns are autocatalytic genetic elements carrying a ribozyme domain responsible for the intron self-splicing reaction, and occasionally a homing endonuclease gene, encoding an endonuclease protein directly involved in the intron mobility at the DNA level. In the genus Naegleria, the group I introns almost all carry a homing endonuclease gene. It was demonstrated that the group I intron in the SSU rDNA of Naegleria spp. was acquired in an ancestral stage and inherited vertically, while it was lost in certain species. The group I introns in the LSU rDNA of Naegleria spp. would be recent horizontal transfers.
SAN2, an amoeboflagellate related to Stachyamoeba (Baumgartner et al, in preparation) has a group I intron in the SSU rDNA, while the ATCC Stachyamoeba, which is also an amoeboflagellate (Murase and De Jonckheere, in preparation) contrary to the type species Stachyamoeba lipophora, does not carry such a group I intron. The amoeba isolate 6-5F related to the ATCC Stachyamoeba does not have a group I intron either (Murase and De Jonckheere, in preparation).
Pseudomastigamoeba longifilum (AF011462) and Pleurostomum flabellatum (Park et al. 2007) also have a group I intron in the SSUrDNA, while Macropharyngomonas halophila has two (AF011465).
In Acrasis rosea 3 different group I introns are present in the SSU rDNA (AF011458) while strain And 12, which cluster with Acrasis rosea (Lara et al. 2007) has none.
The rDNA of all other genera within the Heterolobosea seems to be devoid of group I introns.
Ecology
It appears that each genus is either present only in seawater (or high salinity water) or freshwater (or soil). It might appear that Stachyamoeba is the exception as the typestrain (S. lipophora) is of freshwater origin, while the ATCC strain comes from seawater. However, there are indications, such as the transformation to flagellates, that the ATCC strain, which is used in phylogenetic analyses, does not belong to the genus Stachyamoeba. However, this will never be known with certainty as the typestrain cannot be analyzed anymore as it was lost at CCAP (Culture Collection of Algae and Protozoa).
While all Vahlkampfia spp. are from freshwater some morphologically described Vahlkampfia spp. were isolated from seawater (Page 1983, Smirnov and Fenchel 1996, Anderson et al. 1997) but their exact identity is not known by lack of molecular analysis. This molecular analysis is necessary because the only molecularly investigated marine Vahlkampfia sp., V. damariscottae, turned out to belong to another genus (Brown and De Jonckheere 1999).
Only for the genus Naegleria there is enough information about the dispersal in the environment. It appears that most species are present on all continents except Antarctica (De Jonckheere 2004). In Antarctica and Arctica Naegleria spp. are present that are only found in cold environments (De Jonckheere 2006b). The pathogenic N. fowleri on the other hand is only present in warm climates or hot waters in temperate climates. Of the majority of the Heterolobosea only one strain of one species is described, so that no information is available about their dispersal.
Discussion of Phylogenetic Relationships
The phylogenetic relationships among the Heterolobosea have been studied using nuclear SSU rDNA. These SSU rDNA trees indicate that:
- The Vahlkampfiidae are polyphyletic.
- Willaertia magna was originally regarded as belonging to the genus Naegleria based essentially on cyst morphology. Molecular phylogeny showed, however, that it is a different genus, but it is most closely related to the Naegleria genus.
- The flagellate Heterolobosea genera Stephanopogon and Percolomonas cluster, and together with the other flagellate Pleurostomum flabellatum, they form a big cluster with the majority of the Vahlkampfiidae which are either amoebae or amoeboflagellates.
- The amoeba Heterolobosea genera do not cluster specifically and form different branches with different amoeboflagellates.
- Acrasis rosea (Acrasida) branches within the Vahlkampfiidae.
- Stachyamoeba (Gruberellidae) branches within the Vahlkampfiidae of a second big cluster including the anaerobic Heterolobosea, as well as the marine Heteramoeba and Phaesiobystra.
- Marine genera do not cluster specifically.
- Macropharyngomonas is at the base of the Heterolobosea and is the only group lacking the typical helix 17_1 in the SSU rDNA secondary structure.
Within the Vahlkampfiidae (especially the genera Naegleria, Tetramitus, Vahlkampfia and Willaertia) the internal transcribed spacers, including the 5.8 S rDNA seem to be more useful in the identification and description of species.
References
Anderson O. R., Rogerson A., Hannah F. Three new limax amoebae isolated from marine surface sediments: Vahlkampfia caledonica n. sp., Saccamoeba marina n. sp., and Hartmannella vacuolata n. sp. J. Euk. Microbiol. 44, 33-42, 1997.
Baumgartner, M., Eberhardt, S., De Jonckheere, J. F., Stetter, K. O. Tetramitus thermacidophilus n. sp., an amoeboflagellate from acidic hot springs. J. Eukaryot. Microbiol. in press.
Brown, S., De Jonckheere, J.F. A reevaluation of the amoeba genus Vahlkampfia based on SSUrDNA sequences. Europ. J. Protistol. 35, 49-54, 1999.
Brown S., De Jonckheere J. F. Isolation of a new vahlkampfiid amoeba from soil: Paravahlkampfia lenta. Europ. J. Protistol. 40, 289- 294, 2004.
Brugerolle, G., Simpson, A. G. B. The flagellar apparatus of the Heteroloboseans. J. Eukaryot. Microbiol. 51, 96-107, 2004.
Clark, C. G., Cross, G. A. M. Small-subunit ribosomal RNA sequence from Naegleria gruberi supports the polyphyletic origin of amoebas. Mol. Biol. Evol. 5, 512-518, 1988.
De Jonckheere, J. F. A century of research on the amoeboflagellate genus Naegleria. Acta Protozool. 41, 309-342, 2002.
De Jonckheere, J. F. Molecular definition and the ubiquity of species in the genus Naegleria. Protist 155, 89-103, 2004.
De Jonckheere J. F. Isolation and molecular identification of vahlkampfiid amoebae from an island (Tenerife, Spain). Acta Protozool. 45, 91-96, 2006a.
De Jonckheere J. F. Isolation and molecular identification of vahlkampfiid amoebae from Arctic and sub-Antarctic regions. Europ. J. Protistol. 42, 115-123, 2006b.
De Jonckheere, J. F., Brown, S. The identification of vahlkampfiid amoebae by ITS sequencing. Protist 156, 89-96, 2005.
El Kadiri G., Joyon L., Pussard M. Pernina chaumonti, n. g., n. sp., a new marine amoeba (Rhizopoda, Heterolobosea). Europ. J. Protistol. 28, 43-50, 1992.
Garstecki T., Brown S., De Jonckheere J. F. Description of Vahlkampfia signyensis n. sp. (Heterolobosea), based on morphological, ultrastructural and molecular characteristics. Europ. J. Protistol. 41, 119-127, 2005.
Lara E., Berney C., Ekelund F., Harms H., Chatzinotas A. Molecular comparison of cultivable protozoa from a pristine and a polycyclic aromatic hydrocarbon polluted site. Soil Biol. Biochem. 39, 139-148, 2007.
Nikolaev S.I., Mylnikov A.P., Berney C., Fahrni J., Pawlowski J., Aleshin V.V., Petrov N.B. Molecular phylogenetic analysis places Percolomonas cosmopolitus within Heterolobosea: evolutionary implications. J. Eukaryot. Microbiol. 51, 575-581, 2004.
Page, F. C. Marine Gymnamoebae. Institute of Terrestrial Ecology Culture Centre of Algae and Protozoa, Cambridge England, 1983.
Page, F. C. A New Key to Freshwater and Soil Gymnamoebae. Freshwater Biological Association, Ambleside, Cumbria, 1988.
Page F. C., Blanton R. L. The Heterolobosea (Sarcodina: Rhizopoda), a new class uniting the Schizopyrenida and the Acrasidae (Acrasida). Protistologica 21, 121-132, 1985.
Park J. S., Simpson A. G. B., Lee W. J., Cho B. C. Ultrastructure and phylogenetic placement within Heterolobosea of the previously unclassified, extremely halophilic heterotrophic flagellate Pleurostomum flabellatum (Ruinen 1938) Protist. 158, 397-413, 2007.
Robinson B.S., Christy P.E., De Jonckheere J.F. Observations on a temporary flagellate stage in Vahlkampfiid amoeba Willaertia magna and its significance for the phylogeny of the related genus Naegleria. Biosystems, 23, 75-86,1989.
Robinson B. S., De Jonckheere J. F., Dobson P. J. New Tetramitus species (Heterolobosea, Vahlkampfiidae) from cold aquatic environments. Europ. J. Protistol. 43, 1-7, 2007.
Smirnov A. V., Fenchel T. Vahlkampfia anaerobica n. sp. and Vannella peregrinia n. sp. (Rhizopoda) – Anaerobic amoebae from a marine sediment. Arch. Protistenk. 147, 189-198, 1996.
Yubuki N., Leander B. S. Ultrastructure and molecular phylogeny of Stephanopogon minuta: an enigmatic microeukaryote from marine interstitial environments. Europ. J. Protistol. In press, 2008.
Title Illustrations

| Scientific Name | Naegleria gruberi |
|---|---|
| Copyright |
©
Johan F. De Jonckheere

|
| Scientific Name | Pleurostomum flabellatum |
|---|---|
| Image Use |
 This media file is licensed under the Creative Commons Attribution-NonCommercial License - Version 3.0. This media file is licensed under the Creative Commons Attribution-NonCommercial License - Version 3.0.
|
| Copyright |
© Alastair Simpson

|
| Scientific Name | Stephanopogon minuta |
|---|---|
| Creator | Image taken by Naoji Yubuki |
| Image Use |
 This media file is licensed under the Creative Commons Attribution-NonCommercial License - Version 3.0. This media file is licensed under the Creative Commons Attribution-NonCommercial License - Version 3.0.
|
| Copyright |
© Brian S. Leander

|
About This Page
The author thanks Fred Opperdoes (Trop Unit, de Duve Institute) for giving the opportunity to continue his research in the Trop Unit, and Patrick Van Der Smissen (Cell Biology Unit, de Duve Institute) for the help in taking pictures.
This page is being developed as part of the Tree of Life Web Project Protist Diversity Workshop, co-sponsored by the Canadian Institute for Advanced Research (CIFAR) program in Integrated Microbial Biodiversity and the Tula Foundation.
Johan F. De Jonckheere

de Duve Institute and Scientific Institute of Public Health, Belgium
Correspondence regarding this page should be directed to Johan F. De Jonckheere at
jdjonckh@ben.vub.ac.be
Page copyright © 2009 Johan F. De Jonckheere
All Rights Reserved.
- First online 21 September 2008
- Content changed 21 September 2008
Citing this page:
De Jonckheere, Johan F. 2008. Heterolobosea . Version 21 September 2008 (under construction). http://tolweb.org/Heterolobosea/96360/2008.09.21 in The Tree of Life Web Project, http://tolweb.org/






 Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site
Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site